Case Study: RNA Extraction Quality with RNK4601 miEASY microRNA Kit
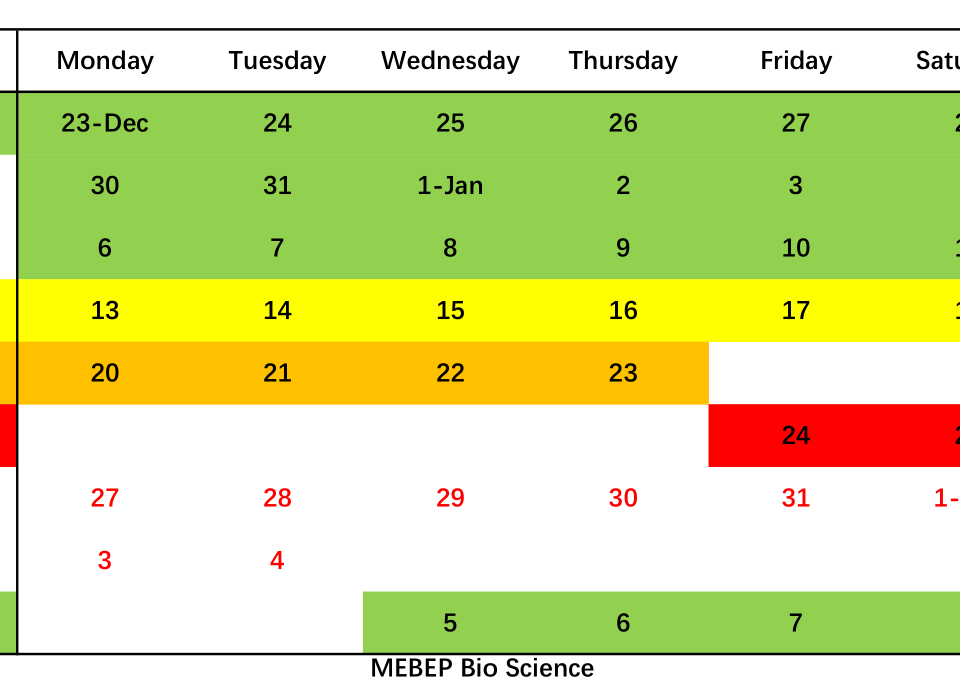
Determine whether RNA is degraded.
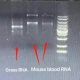
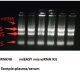
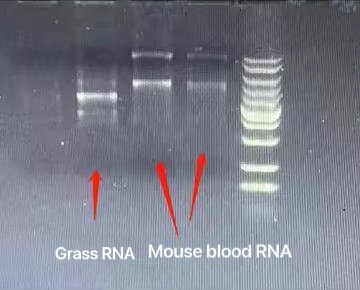
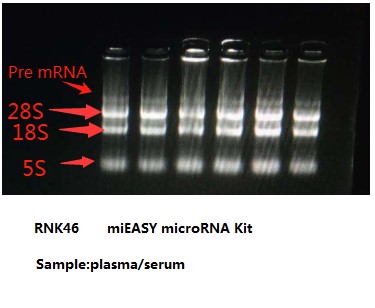
Intact RNA, without degradation, is the most important indicator of RNA extraction quality. It can usually be judged by the ratio of 28S RNA to 18S RNA. The larger the ratio, the higher the integrity. In the above electrophoresis chart, you can see the ratio of 28S RNA to 18S RNA in the RNA extracted by RNK4601. The brightness of the 28S band is greater than the brightness of the 18S band, and the ratio is much greater than 2, indicating that the integrity is very good.
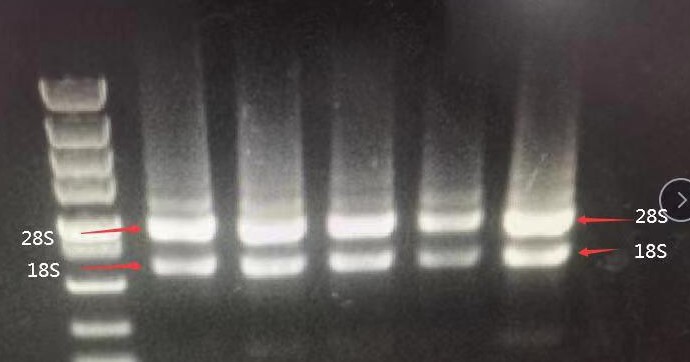
No DNA Residue in the Swimming Lanes
RNK4601 uses a special genome cleaning technology that can quickly clear genomic DNA residues in a short time without DNase digestion or DNase column digestion. It achieves the effect of clearing genomic DNA residues. The resulting RNA can be directly used for downstream fluorescence quantification.
The high molecular weight tailing on the upward sample well in lane 28S is mRNA, which is the messenger RNA. There are hundreds or thousands of large but trace amounts of mRNA. The comprehensive electrophoresis effect is the upper and lower smear bands around lanes 28S and 18S, and the smear background. Fluorescence indicates this is not DNA residue. Therefore, it can also be seen in the electrophoresis chart that there is no DNA residue.
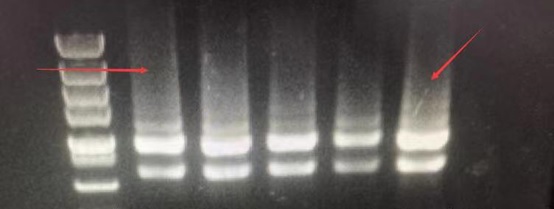
The arrow points to the precursor RNA before shearing from the nucleus. This is not a DNA residue, but the RNA precursor before shearing and maturation. The molecular weight is larger, so it is above 28S. The difference from genomic DNA residues can be determined by the fragment size. Sheared precursor RNA is generally at 6kb, and genomic DNA residues are generally at 10-15kb, with the fragments being obviously larger. This can be verified through RNA digestion experiments. After digestion with RNase (RA02), the fragment that disappears is the precursor RNA, and the fragment that does not disappear is the residual DNA.
Precursor RNA amount is very small, sometimes you can see it, sometimes you can't, it's normal. If it can be seen, because the precursor RNA amount is large, then it can be seen, and not degraded. From another perspective, this proves that the integrity of RNA is very good. The same principle applies: the larger the RNA fragment, the earlier and the easier it is to degrade. If the largest precursor RNA is not degraded, it can basically confirm that the overall RNA integrity is good.
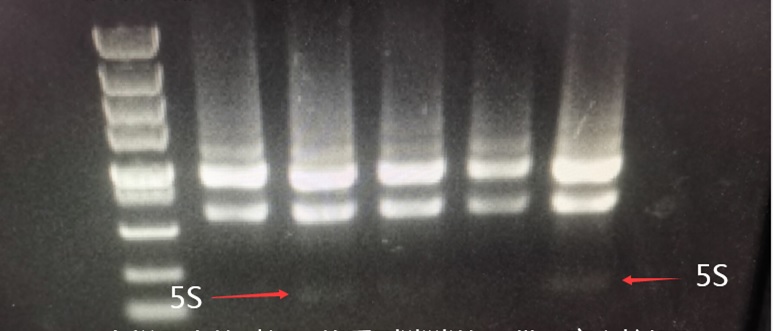
In ordinary RNA extraction kits, the spin column does not adsorb small fragments of nucleic acid, so the small 5S RNA fragment does not adsorb or rarely adsorbs. Therefore, when the spin column extracts RNA and runs electrophoresis, the 5S RNA is invisible or very faint. This is normal.
When loading a large amount of sample, you can see a faint 5S band. When extracting RNA from a spin column, if a very bright band appears at the 5S position, it is often the RNA that has been degraded into small fragments running at this 5S position, indicating degradation.